July 30, 2018 / by Giorgia Cantisani / BioCNN
Transfer Learning and Data Augmentation for Semantic Segmentation in Histopathology
Semantic segmentation means understanding an image at pixel level, i.e. assigning each pixel in the image to an object class. The segmentation of Region Of Interests (ROIs) in high-resolution histopathological images is a task with high clinical relevance normally performed by pathologist however there are some intrinsic problems. In fact it is:
- a time-expensive process
- an user-dependent analysis
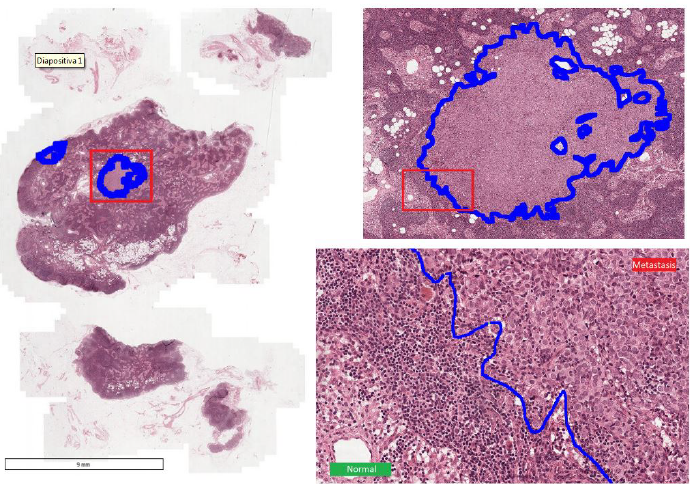
Its automation is desirable as it would hugely reduce the workload of the clinicians, while at the same time reducing the subjectivity of the diagnosis. The segmentation of ROIs can be expressed as a Semantic Segmentation problem for which Deep Learning models are proven to be particular suitable. However there are many difficulties to overcome:
- need for large and fully-annotated datasets for training the models;
- high computational load due to the high resolution of this type of images;
- high variability in the data and in the preprocessing pipelines (mainly due to different cell types, cell densities, stains, magnification levels, and so on).
Proposed approach
To overcome this limitation, we propose to use Transfer Learning and Data Augmentation as possible solutions:
- TRANSFER LEARNING: extracting the knowledge from one source task and apply the learned knowledge to a novel target task. In particular, we fine-tuned the weights of a network trained for tumor segmentation and we fine-tuned it on two novel asks: glands and membrane segmentation.
- DATA AUGMENTATION: increase artificially the number of training samples by a set of transformations. Histological images have no preferential direction and tissues are subjected to elastic deformations. Therefore, we applied random distortions, rotations and flip left right/flip top bottom to simulate these typical variations.
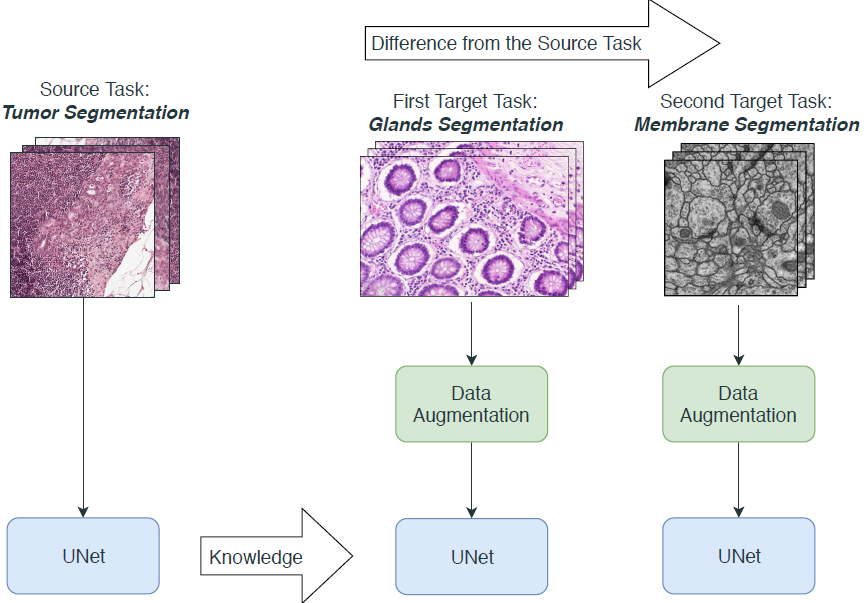
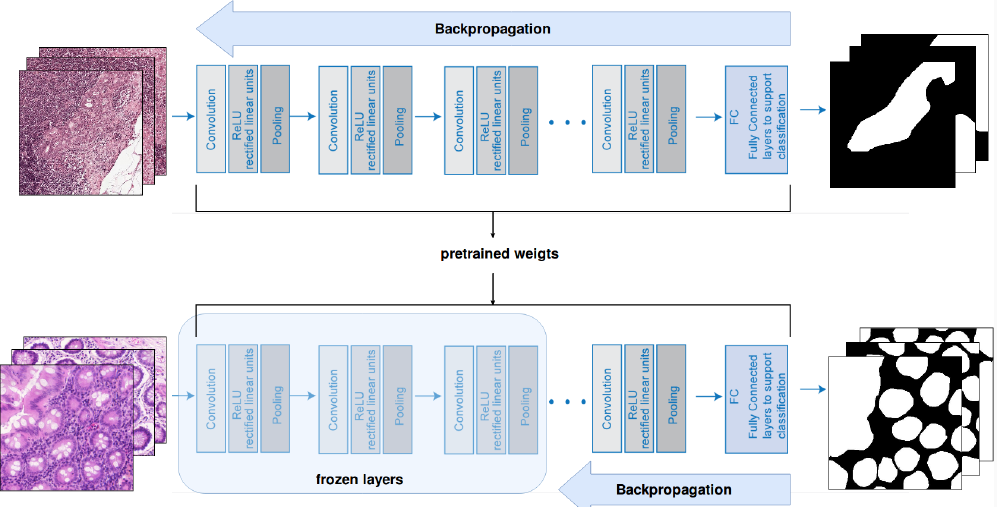
We chose to use UNet, a CNN that works end-to-end, i.e. it outputs directly a segmentation mask starting from the raw image. Moreover, UNet can take as input images of any size because has no FC layers, which is desiderable when using multiple datasets.
Data
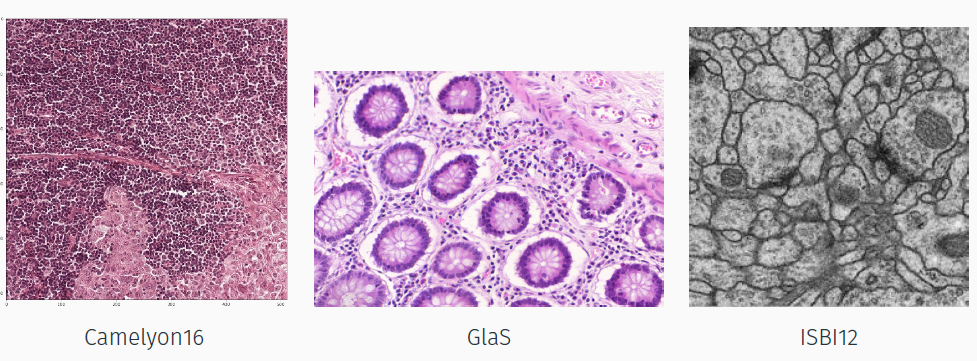
- Camelyon16: 400 H&E lymph node WSIs which can be tiled in order to obtain thousand of cropped images (57048). The task is segmenting cancerous ROIs.
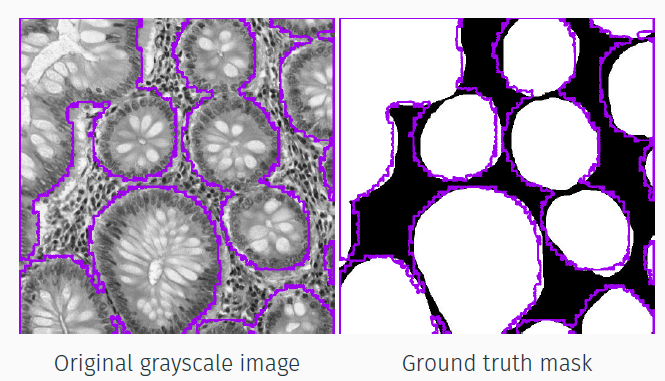
- GlaS: 165 H&E 2D glands sections (content different but the same type of image with respect to Camelyon16). The task is segmenting glands;
- ISBI12: 30 ssTEM images of Drosophila nervous tissue (content and image type different from Camelyon16). The task is segmenting neuron membranes.
Results
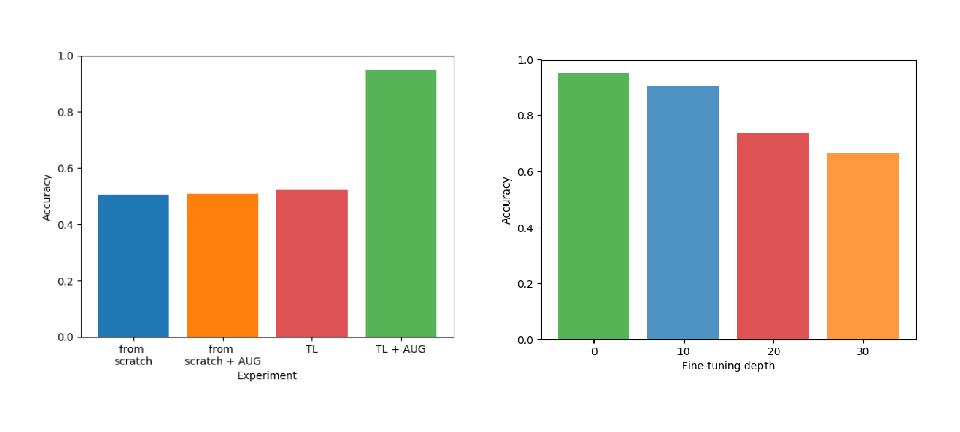
The results show how Transfer Learning and Data Augmentation represent a good solution in the semantic segmentation field:
- The transfer of knowledge is successful when the task are similar. The boundary segmentation task is too different to achieve good performances.
- When the number of frozen layers increases, the performance decreases: deeper features are too specific to the source task.
- Data augmentation always improves accuracy.
Moreover, the required computational resources are acceptable. The best model required no more than 25 GB virtual memory an 8 days CPU-time effectively spent for training.
For more details, please refer to my Master thesis Transfer Learning and Data Augmentation for Semantic Segmentation in Histopathology.
This project was part of my Master Thesis conducted at the Polytechnic of Turin under the supervision of Professor Elisa Ficarra, Santa Di Cataldo, and Francesco Ponzio.